Investigator: Donald M. Anderson (Woods Hole Oceanographic Institution)
Funding Period: October 2005 - September 2009
Partners: Stephen S. Bates (Fisheries and Oceans Canada), Deana L. Erdner (University of Texas Marine Science Institute)
Region: Northeast U.S.
Abstract: The toxic diatom Pseudo-nitzschia produces domoic acid (DA), the cause of amnesic shellfish poisoning. It occurs commonly on the U.S. west coast, and is found in most other coastal regions. In the Gulf of Maine, multiple Pseudo-nitzschia species occur, and some are toxic, but we know nothing of their distribution, nor do we have molecular tools and assays like those used to study west coast DA events. None of the northeastern coastal states routinely monitors for DA in shellfish, so there is the potential for serious human health and ecosystem impacts from Pseudo-nitzschia. This is the first study of its type in this region, and is intended to develop the tools and local expertise to respond to outbreaks, and provide an assessment of the risk from Pseudo-nitzschia.
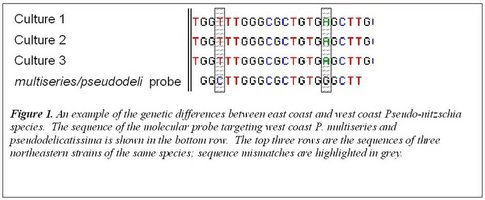 In our first two years, we have isolated Pseudo-nitzschia from across the region and established > 40 cultures. They include four different genetic types, and most are genetically different from the same species found on the west coast. We are currently testing for culture toxicity using multiple methods. We have also preserved samples for analysis of Pseudo-nitzschia, during large-scale research cruises in 2006 and 2007. During both cruises, microscope observations found high numbers of Pseudo-nitzschia cells in the Georges Bank region, suggesting that this area may host a recurrent summer population of Pseudo-nitzschia. This population seems to be present even when high numbers of cells are not observed inshore. Persistent toxicity has been detected in scallop viscera from this same region, in a separate project by PI Anderson. These findings have significant implications for the management of offshore shellfisheries resources. The information from this project will be of direct interest to the shellfish harvesting industry, to coastal managers responsible for public health and seafood safety, as well as those responsible for ecosystem management.
In our first two years, we have isolated Pseudo-nitzschia from across the region and established > 40 cultures. They include four different genetic types, and most are genetically different from the same species found on the west coast. We are currently testing for culture toxicity using multiple methods. We have also preserved samples for analysis of Pseudo-nitzschia, during large-scale research cruises in 2006 and 2007. During both cruises, microscope observations found high numbers of Pseudo-nitzschia cells in the Georges Bank region, suggesting that this area may host a recurrent summer population of Pseudo-nitzschia. This population seems to be present even when high numbers of cells are not observed inshore. Persistent toxicity has been detected in scallop viscera from this same region, in a separate project by PI Anderson. These findings have significant implications for the management of offshore shellfisheries resources. The information from this project will be of direct interest to the shellfish harvesting industry, to coastal managers responsible for public health and seafood safety, as well as those responsible for ecosystem management.Products and Services:
-
Current products and services:
- Tools: a collection of Pseudo-nitzschia cultures from the northeast will be available when they have been identified to species level and tested for toxicity; user - scientists
- Technologies: The ribosomal DNA sequences of our isolates will be deposited in the NCBI public sequence database when they are published; user - scientists
- Characterization: Data collected during the research cruises was made publicly available through http://science.whoi.edu/users/mcgillic/en437/; users - scientists and managers.
- Technologies: Molecular probes and assays for toxic northeastern Pseudo-nitzschia strains will be developed during the project; user - scientists and managers.
- Characterization: Information on the distribution of Pseudo-nitzschia throughout the region will be determined from cruise samples; user - scientists and managers.
- Technologies: This project will assess the threat from Pseudo-nitzschia, and will aid in management decisions regarding the need for monitoring and testing; user - managers.
Proposed products and services:
Investigator: Robert R. Bidigare (PI) - University of Hawaii
Funding Period: September 2004 - August 2008
Partners: Thomas K. Hemscheidt (Associate PI) - University of Hawaii; Angel. A. Yanagihara (Associate PI) - University of Hawaii; E. J. Mathur (Partner) - Diversa Corporation
Region: Pacific Islands
Abstract: The marine biosphere is one of the Earth's richest, but least characterized, habitats. It is home to a large number of diverse marine organisms that are found in environments ranging from temperate to extreme. Biodiversity is now widely accepted as a source for biotechnological discovery and innovation. The application of biotechnology to the marine biosphere can provide new drugs and processes that serve a broad array of sectors, including human health, homeland security and the environment.
 The Hawaiian archipelago is a geographically isolated chain of oceanic islands with high biodiversity and endemism, and is considered to be an environmental 'hot spot' in terms of its rich and often unique animal, plant, and microbial life forms. This unique microbial biodiversity represents a promising resource for the discovery of novel bioactive molecules. Over the last two years, we have greatly improved the efficiency and reproducibility of our anti-microbial screening protocols. Improvements include: (1) stabilization of input to the screening pipeline, (2) elimination of the extraction 'bottleneck,' and (3) optimization of the anti-microbial assays. An example of the screening output is shown in Figure 1. Of the ~900 extracts screened to date, 8% are active against E. coli, 25% are active against vancomycin-resistant enterococci, 31% are active against S. aureus, 34% are active against methicillin-resistant, and 14% are active against the fungus C. albicans. Several bioactive compounds have been isolated and structural determinations are in progress.
The Hawaiian archipelago is a geographically isolated chain of oceanic islands with high biodiversity and endemism, and is considered to be an environmental 'hot spot' in terms of its rich and often unique animal, plant, and microbial life forms. This unique microbial biodiversity represents a promising resource for the discovery of novel bioactive molecules. Over the last two years, we have greatly improved the efficiency and reproducibility of our anti-microbial screening protocols. Improvements include: (1) stabilization of input to the screening pipeline, (2) elimination of the extraction 'bottleneck,' and (3) optimization of the anti-microbial assays. An example of the screening output is shown in Figure 1. Of the ~900 extracts screened to date, 8% are active against E. coli, 25% are active against vancomycin-resistant enterococci, 31% are active against S. aureus, 34% are active against methicillin-resistant, and 14% are active against the fungus C. albicans. Several bioactive compounds have been isolated and structural determinations are in progress.
Products and Services:
- Addition by OHH scientists of 750 new cultures of marine microbes to the CMMED collection
- To date, 482 of these 750 cultures have been subjected to initial screening against a battery of microbial pathogens and cellular and molecular anti cancer assays, with the follow results:
Cultures generating extracts effective against:
Candida albicans 152 Escherichia coli (ATCC 25922) 66 Staphylococcus aureus (ATCC 25922) 154 Multiply Resistant S. aureus (ATCC 43300) 169 Vancomycin Resistant Enterococcus faecium (ATCC 700221) 99 Human Adenocarcinoma Cell Line (SK0V-3) 64 Protein Kinase C (PKC) 7 Mitogen Activated Protein Kinase (MAP Kinase) 2 - Active extracts subjected to bioactivity-guided fractionation: 25
- Compounds purified and characterized: 35
- Novel compounds: 9
- Development of analytical methods for the qualitative and quantitative analysis of .-methylaminoalanine (BMAA) in microbial, plant, and animal tissues (Cox et al. 2005).
- Identification of BMAA in marine cyanobacteria from Hawaiian waters.
- Initial studies of food chain bioaccumulation of BMAA.
- Initial studies of accumulation of BMAA and BMAA metabolites in neural tissues of animals.
- Access to the 3,000 cultures of marine and freshwater microbes in the collection of the Center for Marine Microbiology, Ecology, and Diversity (CMMED).
Investigator: Alexandria Boehm (PI) (Stanford University)
Funding Period: September 2004 - August 2008
Partners: Oliver Fringer (coPI), Kevin Arrigo (coPI), Gary Schoolnik (coPI), Sharyl Rabinovici (collaborator) . Univ. CA Berkeley, Alyson Santoro (PhD student), Yuanan Hu (PhD Student), Sarah Macumber (Masters student), Michael Miller (post-doc), Daniel Keymer (PhD student), Blythe Layton (PhD student), Gert van Dijken (Research Associate), Kevan Yamahara (PhD student). [All affiliations are Stanford University, unless otherwise noted.]
Region: The California Coast, Western NOAA region
Abstract: The goal of our project is to develop early warning systems for beach management and water safety for microbial pollutants.
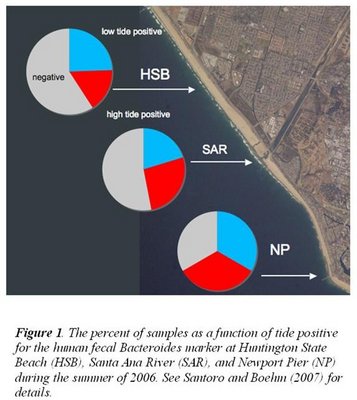 In order to construct early warning systems, it is essential to understand the sources, fate, and transport of target microbial pollutants. In our case, these target microbial pollutants include fecal indicator bacteria, human fecal marker in Bacteroides, chlorophyll a (a measure of dinoflagellate Lingulodinium polyedrum densities that adversely impacted tourism and recreation), and Vibrio cholerae. We are particularly interested in understand how the physics and chemistry of the coastal ocean influence concentrations of these target organisms. We are generating new data for NOAA and also making use of the archived, publicly available data available though NBDC and SCCOOS. The information we are generating is particular important to society who values knowing water quality and understanding how natural and anthropogenic factors influence it. The information we generate is being used by coastal managers all along the California Coast, as well as by researchers in other locations dealing with similar societal concerns.
In order to construct early warning systems, it is essential to understand the sources, fate, and transport of target microbial pollutants. In our case, these target microbial pollutants include fecal indicator bacteria, human fecal marker in Bacteroides, chlorophyll a (a measure of dinoflagellate Lingulodinium polyedrum densities that adversely impacted tourism and recreation), and Vibrio cholerae. We are particularly interested in understand how the physics and chemistry of the coastal ocean influence concentrations of these target organisms. We are generating new data for NOAA and also making use of the archived, publicly available data available though NBDC and SCCOOS. The information we are generating is particular important to society who values knowing water quality and understanding how natural and anthropogenic factors influence it. The information we generate is being used by coastal managers all along the California Coast, as well as by researchers in other locations dealing with similar societal concerns.
We conducted three summers of field work along the California coast to understand dynamics of fecal pollution and chla. Using historical data, we have investigated temporal-spatial patterns in fecal indicator concentrations along the shore, the ability to use environmental factors to model bacterial densities along the coast, and conducted two cost-benefit analyses of different policy scenarios. We conducted mesocosm experiments to examine the effect of grazing on enterococci decline in marine waters. Multi-loci sequence analysis and ERIC-PCR are being used to investigate the diversity and evolutionary history of hundreds of V. cholerae isolates. We are using historical records of incidence of various waterborne illness in Orange County, CA to investigate how environmental factors influence illness. We are studying sand as a source of microbial pollutants and pathogens to coastal waters. Fig. 1 shows our findings of the human fecal Bacteroides marker along the entire coast we studied, and that it does not vary with tide in the same manner as fecal indicator organisms (Santoro and Boehm 2007).
Products and Services:
- Development of molecular tools for investigating source of enterococci and evolutionary history of V. cholerae.
- models to predict when microbial pollution levels will be greatest based on environmental factors
- model to study fate of a wastewater plume
- data collection of biological and physical oceanography spanning numerous years
- model of how environmental factors influence illness rates of local population
- application of technology to coastal resource management through synthesis, integration, training, and the development of new management tools
- characterization of the California Coast in terms of water quality and physical oceanography
Investigators: Greg Boyer, Professor of Biochemistry (State University of New York, College of Environmental Science and Forestry), Steven Wilhelm, Associate Professor of Microbiology (The University of Tennessee)
Funding Period: October 2005 - September 2009
Partners: NOAA's GLERL laboratory, NOAA's MERHAB-LGL
Region: lower Laurentian Great Lakes: Lake Erie, Lake Ontario and Lake Huron, and their associated watersheds.
Abstract: Freshwater biotoxins are markedly understudied relative to those in marine systems and only microcystins are currently addressed to any significant degree.
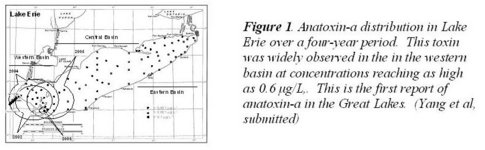 Recent evidence indicates that new biotoxins and toxic strain new to the Great Lakes (cylindrospermopsin, anatoxin-a, Planktothrix) may be present. Supporting and expanding upon NOAA GLERL's efforts with Microcystis and microcystins, we collected and characterized novel toxigenic cyanobacteria from the Laurentian Great Lakes as well as other freshwater systems, establish molecular and biochemical diagnostics for their presence and the presence of these specific toxins. It addresses questions such as .Who are the major producers of the microcystins in Lake Erie and what is the contribution by species other than Microcystis to the total microcystin burden found in the lake?. and .What in the contribution of cyanobacterial neurotoxins to human health risks from cyanobacterial blooms in Lake Erie..
Recent evidence indicates that new biotoxins and toxic strain new to the Great Lakes (cylindrospermopsin, anatoxin-a, Planktothrix) may be present. Supporting and expanding upon NOAA GLERL's efforts with Microcystis and microcystins, we collected and characterized novel toxigenic cyanobacteria from the Laurentian Great Lakes as well as other freshwater systems, establish molecular and biochemical diagnostics for their presence and the presence of these specific toxins. It addresses questions such as .Who are the major producers of the microcystins in Lake Erie and what is the contribution by species other than Microcystis to the total microcystin burden found in the lake?. and .What in the contribution of cyanobacterial neurotoxins to human health risks from cyanobacterial blooms in Lake Erie..
Investigator: Sylvain De Guise (PI) . University of Connecticut; Milton Levin (post-doc) . University of Connecticut
Funding Period: September 2004 - August 2008
Region: North Atlantic
Abstract: The oceans and their resources have become increasingly well recognized as sources of food for humans and for the biodiversity they harbor. Nevertheless, the widespread dissemination of pollutants in the aquatic food chain has prompted concerns on the possible health risks associated with consumption of seafood, as exemplified by the recent controversy over polychlorinated biphenyls (PCBs) in aquaculture raised salmon.
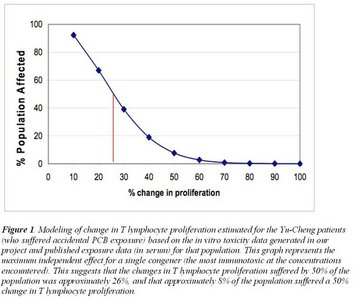 Risk assessments for PCBs are largely based on the dioxin toxic equivalency (TEQ) approach, which assumes that toxicity is exerted through binding of the dioxin receptor. Therefore, the dioxin-like (non-ortho and mono-ortho) PCB congeners that have the conformation to bind this receptor are considered .toxic., while those which cannot (di-ortho PCBs) are considered safe. We tested the toxicity of PCBs from these different classes on immune functions (T- and B-lymphocyte proliferation, neutrophil and monocyte phagocytosis and respiratory burst) of several species of marine mammals, humans and mice to determine if a structure-activity relationship exists across species. Results to date suggest that the dose-response curves within and between .ortho. classes for a given species and cell type often showed significant differences. Furthermore, cluster analysis showed that consistent structure-activity relationships could not be determined based on .ortho. classification. Overall, di-ortho congeners (traditionally considered relatively safe) generally produced greater immunomodulatory effects than the non-ortho (dioxin-like) congeners. This work suggests that the TEQ approach to risk assessment is likely to miscalculate the immunomodulatory potential of PCBs. Our results were used in a preliminary, species-specific, mechanistically-based modeling exercise to assess and quantify the risk of immunotoxic effects at the population level given existing exposure data. Such data will be important for the development of science-based policies to assure long-term public and environmental health in view of global changes in our oceans.
Risk assessments for PCBs are largely based on the dioxin toxic equivalency (TEQ) approach, which assumes that toxicity is exerted through binding of the dioxin receptor. Therefore, the dioxin-like (non-ortho and mono-ortho) PCB congeners that have the conformation to bind this receptor are considered .toxic., while those which cannot (di-ortho PCBs) are considered safe. We tested the toxicity of PCBs from these different classes on immune functions (T- and B-lymphocyte proliferation, neutrophil and monocyte phagocytosis and respiratory burst) of several species of marine mammals, humans and mice to determine if a structure-activity relationship exists across species. Results to date suggest that the dose-response curves within and between .ortho. classes for a given species and cell type often showed significant differences. Furthermore, cluster analysis showed that consistent structure-activity relationships could not be determined based on .ortho. classification. Overall, di-ortho congeners (traditionally considered relatively safe) generally produced greater immunomodulatory effects than the non-ortho (dioxin-like) congeners. This work suggests that the TEQ approach to risk assessment is likely to miscalculate the immunomodulatory potential of PCBs. Our results were used in a preliminary, species-specific, mechanistically-based modeling exercise to assess and quantify the risk of immunotoxic effects at the population level given existing exposure data. Such data will be important for the development of science-based policies to assure long-term public and environmental health in view of global changes in our oceans.
Products and Services:
- New tool: determination of the relative immunotoxicity of PCBs in different species, with the confirmation that the structure of PCBs cannot predict their immunotoxicity, and that the mouse model cannot predict toxicity in other species.
- New tool: a new species-specific, mechanistically-based model to quantify the risk of immunotoxicity at the population level, given data on exposure level, as the basis for the development of science-based policies and informed risk management to assure long-term public and environmental health in view of global changes in our oceans.
Investigator: J. Lawrence Dunn (Mystic Aquarium)
Funding Period: September 2004 - August 2008)
Region: Analysis of samples from all coastal Atlantic and Pacific NOAA regions as well as the Canadian Arctic
Abstract: Brucella, a pathogen known for centuries due to its devastating health and reproductive effects in humans and livestock, has recently been isolated from marine mammals. While the economic effects of brucellosis in the terrestrial environment have been staggering, the effects of marine origin Brucella are unquantified. Our efforts center on the development of diagnostics with increased sensitivity, specificity and efficiency, while concurrently improving our understanding of the demographics and pathobiology of infection in humans and marine mammals.
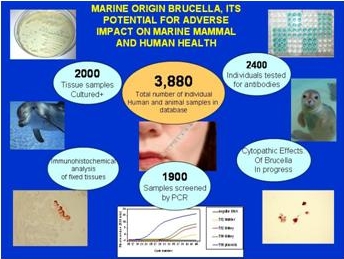 Published reports and our analyses suggest that in cetaceans marine origin Brucella infection can decrease conception rates and increase spontaneous abortion and stillbirth rates. Though evidence of exposure is high in many seal/sea lion populations, until now no studies have documented significant pathologic changes in this group. We recently documented an association between Brucella isolation or its molecular detection and abortions or premature delivery in California sea lions. Marine origin Brucella also poses a potential threat to humans who come into close contact with marine mammal body fluids and tissues. Thus far, four cases of marine origin brucellosis in humans have been reported in the literature. None of the eighty one marine mammal researchers and animal care personnel tested utilizing our assay showed antibodies to Brucella. This fall, in concert with collaborators at McGill University School of Medicine, we will begin analysis on several hundred serum samples to determine the antibody levels to Brucella in subsistence hunting Inuit peoples of Nunavut, a population with greater exposure to this zoonotic pathogen. Our improved diagnostics have been utilized by the U.S. Navy, NOAA Fisheries, collaborating researchers at numerous universities and public aquariums. Upon publication of the protocols employed in the diagnostics they will be available to any group interested in developing a similar capability.
Published reports and our analyses suggest that in cetaceans marine origin Brucella infection can decrease conception rates and increase spontaneous abortion and stillbirth rates. Though evidence of exposure is high in many seal/sea lion populations, until now no studies have documented significant pathologic changes in this group. We recently documented an association between Brucella isolation or its molecular detection and abortions or premature delivery in California sea lions. Marine origin Brucella also poses a potential threat to humans who come into close contact with marine mammal body fluids and tissues. Thus far, four cases of marine origin brucellosis in humans have been reported in the literature. None of the eighty one marine mammal researchers and animal care personnel tested utilizing our assay showed antibodies to Brucella. This fall, in concert with collaborators at McGill University School of Medicine, we will begin analysis on several hundred serum samples to determine the antibody levels to Brucella in subsistence hunting Inuit peoples of Nunavut, a population with greater exposure to this zoonotic pathogen. Our improved diagnostics have been utilized by the U.S. Navy, NOAA Fisheries, collaborating researchers at numerous universities and public aquariums. Upon publication of the protocols employed in the diagnostics they will be available to any group interested in developing a similar capability.
Products and Services:
- Triplex real-time PCR assay . Sensitivity testing of our refined molecular assay has shown a 95% detection limit of 20 fg of bacterial DNA, or approximately 5 bacteria.
- Immunohistochemical detection of Brucella in fixed tissues using a horseradish peroxidase-based detection system on tissue sections.
- cELISA serologic assay . Optimized for detection of antibodies to marine and terrestrial Brucella in serum of any species.
- iELISA serologic assay . Optimized for detection of antibodies to Brucella in bottlenose dolphins.
- Microbiologic culture of tissues and fluids . optimized for growth of Brucella species.
Investigators: Evan Gallagher, University of Washington
Funding Period: October 2005 - September 2009
Partners: Dr. Tracy Collier, Ph.D., National Marine Fisheries Service/NOAA, Seattle, Washington; Federico M. Farin, M.D. Director, Functional Genomics Laboratory, University of Washington; Sandra O'Neill, Research Scientist, Washington Department of Fisheries and Wildlife; Heather Stapleton, Ph.D., Duke University
Region: Puget Sound region, Pacific Northwest United States
Abstract: The polybrominated diphenyl ethers (PBDEs) are a class of flame retardants that include congeners whose tissue residues in resident Puget Sound Chinook salmon are high relative to other salmonids. BDE 47 (2,2.,4,4.-tetraBDE) is one of the more prevalent BDE congeners in Chinook tissues, as well as in most fish and wildlife samples, this despite the fact that BDE 47 is not the congener produced commercially in greatest mass.
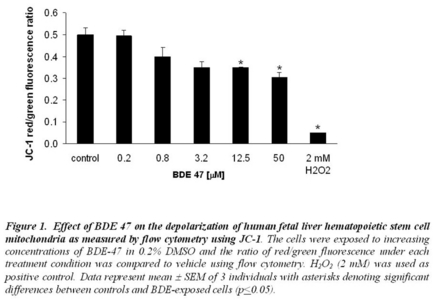 Thus, the origins of the high levels of BDE 47 in resident Chinook and the mechanisms of PBDE-mediated toxicity are poorly understood. Particularly problematic is the lack of knowledge regarding; 1) structure-activity relationships of BDE congeners, 2) mixture effects resulting from the real-world scenarios of exposures to multiple compounds, and 3) human in utero cell toxicity resulting from consumption of contaminated fish during pregnancy. Preliminary in vitro metabolism studies suggest that the origin of the high concentrations of BDE 47 in Chinook tissues may not be due to metabolic debromination of higher molecular weight congeners such as BDE 99 or 209. Studies in zebrafish embryos and larvae using the predominant BDE congeners found in resident Chinook (BDEs 47, 49, 99 and 100) revealed several phenotypic effects of developmental exposure as well as similarities and differences in these endpoints between congeners. Comparative flow cytometry studies of BDE 47 cell injury using primary hematopoietic cells isolated from human fetal liver and also using salmonid cells reveal that mitochondrial injury and cellular oxidative stress are important mechanisms of BDE 47 toxicity, and that these sublethal endpoints may be useful in assessing the toxicity of PBDEs. However, kinetic modeling of BDE 47 cellular uptake and recently available human fetal tissue residue data suggest that the observed cell toxicity may be of less relevance in utero. Ongoing microarray studies using biomedical models (e.g. zebrafish and human fetal stem cells) are directed towards identifying other potential mechanisms of PBDE injury relevant to humans. The results of our ongoing studies are discussed in the context of a model Oceans-Human Health interaction and reducing the uncertainty of potential human risks associated with consumption of Puget Sound Chinook.
Thus, the origins of the high levels of BDE 47 in resident Chinook and the mechanisms of PBDE-mediated toxicity are poorly understood. Particularly problematic is the lack of knowledge regarding; 1) structure-activity relationships of BDE congeners, 2) mixture effects resulting from the real-world scenarios of exposures to multiple compounds, and 3) human in utero cell toxicity resulting from consumption of contaminated fish during pregnancy. Preliminary in vitro metabolism studies suggest that the origin of the high concentrations of BDE 47 in Chinook tissues may not be due to metabolic debromination of higher molecular weight congeners such as BDE 99 or 209. Studies in zebrafish embryos and larvae using the predominant BDE congeners found in resident Chinook (BDEs 47, 49, 99 and 100) revealed several phenotypic effects of developmental exposure as well as similarities and differences in these endpoints between congeners. Comparative flow cytometry studies of BDE 47 cell injury using primary hematopoietic cells isolated from human fetal liver and also using salmonid cells reveal that mitochondrial injury and cellular oxidative stress are important mechanisms of BDE 47 toxicity, and that these sublethal endpoints may be useful in assessing the toxicity of PBDEs. However, kinetic modeling of BDE 47 cellular uptake and recently available human fetal tissue residue data suggest that the observed cell toxicity may be of less relevance in utero. Ongoing microarray studies using biomedical models (e.g. zebrafish and human fetal stem cells) are directed towards identifying other potential mechanisms of PBDE injury relevant to humans. The results of our ongoing studies are discussed in the context of a model Oceans-Human Health interaction and reducing the uncertainty of potential human risks associated with consumption of Puget Sound Chinook.
Products and Services:
- Human fetal liver derived hematopoietic stem cells are now validated as a research tool that is highly relevant for in utero toxicology studies
- Anticipated generation of microarray databases in zebrafish and humans that can be used by other researchers involved in PBDE research
- Quantitative PCR assays that are specific for Chinook salmon and that can be used by other salmon researchers, including population genetics, physiologist, biochemists, toxicologists to measure gene expression.
Investigator: D. Jay Grimes (University of Southern Mississippi)
Funding Period: September 2004 - August 2008
Region: This study focuses on the northern Gulf of Mexico, specifically on oyster reefs along coastal Mississippi and Alabama.
Abstract:
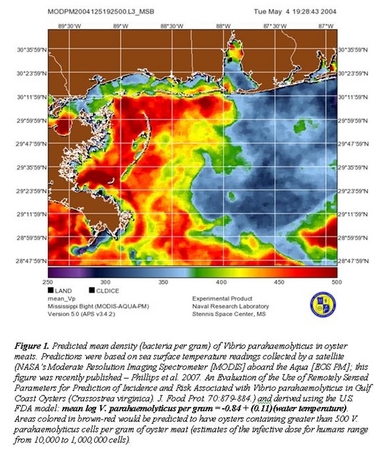 The NOAA OHHI-funded project entitled "The Use of Remote Sensing and Molecular Detection to Predict the Risk of Infection by Vibrio parahaemolyticus" examines the relationship between environmental parameters and the densities of an ocean-dwelling human pathogen, V. parahaemolyticus. Parameters include sea surface temperature (SST), turbidity, salinity, and chlorophyll, and they are measured both in situ (in the field) and remotely (using MODIS satellite-based remote sensing). Parameters are correlated with the densities of Vp-specific genes indicative of total Vp and hemolysin-carrying pathogenic Vp. These correlations are being used to identify environmental signatures that can predict when Vp levels in oysters and overlying waters in the northern Gulf of Mexico reach dangerous levels. In the latest phase of the study (2006-2007), total Vp densities in oysters have correlated significantly with SST, chlorophyll, and turbidity, and total Vp densities in overlying waters have correlated significantly with SST and partially with turbidity. No significant correlations have been identified for pathogenic Vp, likely because these represent such a small proportion of the total Vp population. In addition to Vp, data on a similar ocean-dwelling human pathogen, V. vulnificus (Vv), has been collected in a parallel study, and similar correlations have been identified: SST, chlorophyll, and turbidity in oysters, and SST in overlying waters. Furthermore, quantitation of phytoplankton and zooplankton has identified correlations between diatoms and both Vp and Vv. Several side projects have been added, included phylogenetic typing of pathogenic Vp found in the environment and examination of the Vp Type III Secretion Systems, for which mutants are currently being generated. This project and side projects address human pathogens found naturally in the ocean and will produce prediction tools that can be used by both regulatory agencies and the public to minimize the human health impacts of Vibrio spp.
The NOAA OHHI-funded project entitled "The Use of Remote Sensing and Molecular Detection to Predict the Risk of Infection by Vibrio parahaemolyticus" examines the relationship between environmental parameters and the densities of an ocean-dwelling human pathogen, V. parahaemolyticus. Parameters include sea surface temperature (SST), turbidity, salinity, and chlorophyll, and they are measured both in situ (in the field) and remotely (using MODIS satellite-based remote sensing). Parameters are correlated with the densities of Vp-specific genes indicative of total Vp and hemolysin-carrying pathogenic Vp. These correlations are being used to identify environmental signatures that can predict when Vp levels in oysters and overlying waters in the northern Gulf of Mexico reach dangerous levels. In the latest phase of the study (2006-2007), total Vp densities in oysters have correlated significantly with SST, chlorophyll, and turbidity, and total Vp densities in overlying waters have correlated significantly with SST and partially with turbidity. No significant correlations have been identified for pathogenic Vp, likely because these represent such a small proportion of the total Vp population. In addition to Vp, data on a similar ocean-dwelling human pathogen, V. vulnificus (Vv), has been collected in a parallel study, and similar correlations have been identified: SST, chlorophyll, and turbidity in oysters, and SST in overlying waters. Furthermore, quantitation of phytoplankton and zooplankton has identified correlations between diatoms and both Vp and Vv. Several side projects have been added, included phylogenetic typing of pathogenic Vp found in the environment and examination of the Vp Type III Secretion Systems, for which mutants are currently being generated. This project and side projects address human pathogens found naturally in the ocean and will produce prediction tools that can be used by both regulatory agencies and the public to minimize the human health impacts of Vibrio spp.
Products and Services:
- Using a tool (remote sensing, RS, by satellite) to gather environmental data that can be used to more accurately and quickly forecast human health risks from consuming raw oysters; these data have been collected for the past three years and continue to be collected on a twice-daily basis.
- Using RS data to produce a risk model (either a new model or an improved FDA model) that will be used by consumers, seafood dealers, fishermen, and environmental managers to forecast (predict) human health risks from consuming raw oysters.
- Outreach efforts to better inform the public (consumers, seafood dealers, fishermen, and environmental managers) about the risks associated with consuming raw oysters.
Investigator: Robin Overstreet (University of Southern Mississippi)
Funding Period: August 2008 - July 2011
Region:
Abstract: Parasites are a real threat to seafood consumers and handlers, even though not generally recognized by the medical community and seafood industry. For several years, helminths in particular have been ignored as a threat because proper freezing or cooking can prevent human infections. Recently, for a variety of reasons, humans are once again being infected and affected by parasitic infections. Reasons for increased risk in the US include new cuisines; increased immigration of people; increased consumption of home-prepared seafood; different sources of seafood products resulting from climate changes, overfished stocks of commonly-eaten products, and increased importation; improved transportation and storing systems; recognized hypersensitivity of individuals handling infected seafood; and high numbers of immuno-compromised individuals resulting from medications and disease. One objective of the project is to provide identifications and relative abundance of collected infectious parasitic agents, primarily from the northern Gulf of Mexico but also from products from other sources available to consumers in the US, including aquaculture products and imports. Another objective is to develop methods and tools necessary to rapidly identify those parasites and determine the potential for representative parasites to harm small mammalian hosts and, hence, perhaps humans. We suspect that we can establish specific parasites to serve as early warning systems to minimize human infections with coastal and marine parasites and forecast the potential threats and long-term risks to human health. The intended benefits and deliverables include disseminating information on potentially harmful and aesthetically displeasing seafood parasites in handbook form to the requesting state, regional, and federal fisheries agencies (e.g., Mississippi Department of Marine Resources [DMR], National Marine Fisheries Service [NMFS], and US Food and Drug Administration [FDA]) plus other interested organizations to use in managing seafood imports, sales, and consumption; dealing with concerned consumers, such as those who contact Overstreet and governmental agencies frequently; and alerting the public health community. Findings also will be published in peer-reviewed, scientific journals.
Products and Services:
Investigator: Frances Gulland (The Marine Mammal Center)
Funding Period: September 2004 - August 2007
Region: California, Western region of NOAA
Abstract:
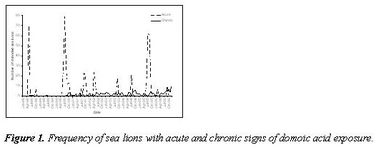 This project has used California sea lions as sentinels of ocean change that can impact human health; specifically it has identified effects of sub-lethal exposure of sea lions to domoic acid in naturally exposed wild populations. By combining epidemiological, clinical and pathology data, domoic acid was associated with the development of epilepsy in sea lions. This syndrome has been increasing, although acute poisoning events also continue. Regular sampling of water in sea lion foraging habitat demonstrated algal blooms in foraging sites, testing of sea lion urine demonstrated domoic acid ingestion, and magnetic resonance imaging, electroencephalography and histopathology showed progressive hippocampal atrophy associated with subclinical seizures in wild sea lions. A secondary effect was also identified: a cardiomyopathy characterized by apoptosis of the conducting system. A third impact of low levels of domoic acid exposure, reproductive failure, was also identified by sampling of fetuses found on sea lion breeding rookeries. Domoic acid was found to account for 25% of abortions, and to cross the placenta of pregnant animals, accumulating in amniotic fluid.
This project has used California sea lions as sentinels of ocean change that can impact human health; specifically it has identified effects of sub-lethal exposure of sea lions to domoic acid in naturally exposed wild populations. By combining epidemiological, clinical and pathology data, domoic acid was associated with the development of epilepsy in sea lions. This syndrome has been increasing, although acute poisoning events also continue. Regular sampling of water in sea lion foraging habitat demonstrated algal blooms in foraging sites, testing of sea lion urine demonstrated domoic acid ingestion, and magnetic resonance imaging, electroencephalography and histopathology showed progressive hippocampal atrophy associated with subclinical seizures in wild sea lions. A secondary effect was also identified: a cardiomyopathy characterized by apoptosis of the conducting system. A third impact of low levels of domoic acid exposure, reproductive failure, was also identified by sampling of fetuses found on sea lion breeding rookeries. Domoic acid was found to account for 25% of abortions, and to cross the placenta of pregnant animals, accumulating in amniotic fluid.
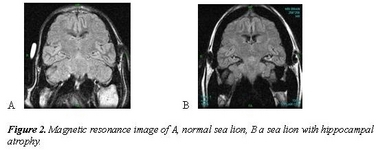 These results have been used by researchers investigating low levels of exposure in mice to guide laboratory studies to further investigate the effects of domoic acid on hippocampal development. They have also been used by policy makes in California to highlight the concern over ocean changes resulting from human activities that may enhance development of harmful algal blooms. This work is important as it suggests chronic health effects of domoic acid exposure can occur in animals eating levels below the level recognized to cause mortality. Furthermore, these effects are observed in California sea lion, animals that share many prey items with humans, such as salmon, squid, hake and rockfish.
These results have been used by researchers investigating low levels of exposure in mice to guide laboratory studies to further investigate the effects of domoic acid on hippocampal development. They have also been used by policy makes in California to highlight the concern over ocean changes resulting from human activities that may enhance development of harmful algal blooms. This work is important as it suggests chronic health effects of domoic acid exposure can occur in animals eating levels below the level recognized to cause mortality. Furthermore, these effects are observed in California sea lion, animals that share many prey items with humans, such as salmon, squid, hake and rockfish.
Products and Services: Increased sampling of coastal sites for domoic acid producing blooms by the California department of Health Services
Investigator: Erin K. Lipp (University of Georgia)
Funding Period: October 2004 - September 2009
Partners: GA Div. of Public Health & University of Georgia; Centers for Disease Control and Prevention
Region: South Atlantic Bight/Coastal Georgia
Abstract: This study is investigating the relationship between climatic variability, estuarine water quality and human health, from direct exposure to pathogens or by consumption of shellfish. Better detection and improved understanding of the fate and ecology of pathogens are critical goals for reducing human exposure. Working with the Georgia Dept. of Natural Resources we are assessing environmental levels and potential exposures through water and estuarine reservoirs of the pathogens Vibrio vulnificus, V. parahaemolyticus, V. cholerae and noroviruses, as well as evaluating traditional and novel water quality indicators.
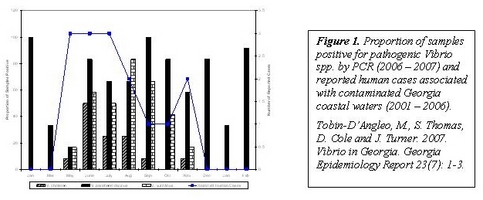 Research to date has shown that while Vibrio illness in Georgia residents is relatively low, 10% of the reported cases could be traced back to Georgia coastal waters and of these 60% were due to V. vulnificus and 20% were due to V. parahaemolyticus. This is consistent with our field results showing that vibrios are readily recovered from Georgia's shellfish harvesting waters, including V. cholerae, V. vulnificus and V. parahaemolyticus, especially when temperatures exceeded 15oC. V. cholerae was detected least frequently among the pathogenic species but 26% did contain the cholera toxin gene. V. vulnificus was detected from 50% of water samples. V. parahaemolyticus was unique in that it was frequently detected at temperatures below 15o and also included virulent strains as evidenced by the detection of multiple virulence gene targets. While Vibrio spp. occur naturally in estuarine water, other enteric pathogens are commonly introduced by improper handling and disposal of sewage. We evaluated the utility of traditional fecal indicator bacteria to predict contamination of these estuaries with enteric viruses and found that enterococci were better associated with the presence of human enteroviruses than were fecal coliform bacteria; however, data indicate that enterococci may grow within estuarine phyto- and/or zooplankton and could represent a niche for environmental persistence that is uncoupled from sewage sources. Given that our research to date indicates possible contamination of these shellfish sources despite acceptable levels of fecal indicator bacteria, we have worked in collaboration with the CDC's calicivirus lab to develop a rapid and streamlined quantitative detection system for noroviruses from shellfish and other estuarine samples. Norovirus are estimated to be the primary cause of viral gastroenteritis associated with shellfish consumption but detection from environmental samples can be laborious and non-quantitative. Our methods are being applied to direct norovirus from these sources. Data on environmental prevalence of Vibrio pathogens and norovirus will be used to develop a risk assessment model for commercial harvesters along Georgia's coast.
Research to date has shown that while Vibrio illness in Georgia residents is relatively low, 10% of the reported cases could be traced back to Georgia coastal waters and of these 60% were due to V. vulnificus and 20% were due to V. parahaemolyticus. This is consistent with our field results showing that vibrios are readily recovered from Georgia's shellfish harvesting waters, including V. cholerae, V. vulnificus and V. parahaemolyticus, especially when temperatures exceeded 15oC. V. cholerae was detected least frequently among the pathogenic species but 26% did contain the cholera toxin gene. V. vulnificus was detected from 50% of water samples. V. parahaemolyticus was unique in that it was frequently detected at temperatures below 15o and also included virulent strains as evidenced by the detection of multiple virulence gene targets. While Vibrio spp. occur naturally in estuarine water, other enteric pathogens are commonly introduced by improper handling and disposal of sewage. We evaluated the utility of traditional fecal indicator bacteria to predict contamination of these estuaries with enteric viruses and found that enterococci were better associated with the presence of human enteroviruses than were fecal coliform bacteria; however, data indicate that enterococci may grow within estuarine phyto- and/or zooplankton and could represent a niche for environmental persistence that is uncoupled from sewage sources. Given that our research to date indicates possible contamination of these shellfish sources despite acceptable levels of fecal indicator bacteria, we have worked in collaboration with the CDC's calicivirus lab to develop a rapid and streamlined quantitative detection system for noroviruses from shellfish and other estuarine samples. Norovirus are estimated to be the primary cause of viral gastroenteritis associated with shellfish consumption but detection from environmental samples can be laborious and non-quantitative. Our methods are being applied to direct norovirus from these sources. Data on environmental prevalence of Vibrio pathogens and norovirus will be used to develop a risk assessment model for commercial harvesters along Georgia's coast.
Products and Services:
- Characterization of Georgia's shellfish harvesting water quality with regard to key pathogens. Information is being shared with the GA Dept. of Natural Resources shellfish and water quality divisions.
- Characterization of the distribution of bacterial and viral pathogens among potential estuarine reservoirs.
- New information on disease outcomes in Georgia's coastal community based on analysis of hospital discharge data in addition to reportable diseases and cases.
- New method for the rapid and quantitative detection of noroviruses from oysters
Investigator: Sandra McLellan (Great Lakes WATER Institute, University of Wisconsin-Milwaukee)
Funding Period: October 2005 - September 2009
Region: Our study site is the nearshore waters (0-20 km) of Lake Michigan adjacent to the Milwaukee Harbor, an area heavily impacted by drainage from the Milwaukee River Basin, which encompasses 850 square miles of rural, suburban and urbanized watersheds.
Abstract:
 Our project is to couple hydrodynamic modeling and particle distribution studies with molecular detection methods to predict pathogen fate in the Great Lakes. Recent findings from our work have shown indicators of human sources of fecal pollution and viruses, including adenovirus, norovirus, enterovirus, and Hepatitis A, are present at detectable levels following rain events that introduce only stormwater, as well as combined sewage into Lake Michigan. Pollution plumes are rapidly diluted once they extend more than 2.5 km offshore, however evidence of fecal pollution can be found using molecular methods as far as 8-10 km into open waters. Fecal indicator bacteria within the plume were found to be attached to small particles ranging from 10-30 . m. Clostridium perfringens, a spore forming gut bacterium, was found to accumulate to high levels in sediment traps within the plume tract, and in traps deployed 1 km offshore of a nearby urban beach (please see figure),
Our project is to couple hydrodynamic modeling and particle distribution studies with molecular detection methods to predict pathogen fate in the Great Lakes. Recent findings from our work have shown indicators of human sources of fecal pollution and viruses, including adenovirus, norovirus, enterovirus, and Hepatitis A, are present at detectable levels following rain events that introduce only stormwater, as well as combined sewage into Lake Michigan. Pollution plumes are rapidly diluted once they extend more than 2.5 km offshore, however evidence of fecal pollution can be found using molecular methods as far as 8-10 km into open waters. Fecal indicator bacteria within the plume were found to be attached to small particles ranging from 10-30 . m. Clostridium perfringens, a spore forming gut bacterium, was found to accumulate to high levels in sediment traps within the plume tract, and in traps deployed 1 km offshore of a nearby urban beach (please see figure),  which demonstrates that sediments may serve as long term reservoirs for commensal bacteria and pathogens. Sediment resuspension was extremely high near the beach site, suggesting beach sites maybe impacted by pathogen reservoirs in sediments. Particle residence time studies using the radionuclides Be-7, Pb-210, and Cs-137 as natural tracers will allow us to correlate sediment dynamics with pathogen persistence. Hydrodynamic modeling efforts have validated a fine grid model at 100 m and 50 m grid resolutions, where both wind vectors and whole lake circulation were found to drive currents and therefore plume dynamics. The open boundary conditions are obtained from the NOAA whole lake circulation model maintained by GLERL. Further modeling work will integrate the particle and biological components. The frequency and intensity of storms is expected to increase in future years, which would correspond to increases in pathogen loads carried by stormwater runoff and combined sewer overflows. This work will be important for water resource managers, regulatory agencies, and urban planners so that they may implement strategies for minimizing pathogen inputs into the Great Lakes.
which demonstrates that sediments may serve as long term reservoirs for commensal bacteria and pathogens. Sediment resuspension was extremely high near the beach site, suggesting beach sites maybe impacted by pathogen reservoirs in sediments. Particle residence time studies using the radionuclides Be-7, Pb-210, and Cs-137 as natural tracers will allow us to correlate sediment dynamics with pathogen persistence. Hydrodynamic modeling efforts have validated a fine grid model at 100 m and 50 m grid resolutions, where both wind vectors and whole lake circulation were found to drive currents and therefore plume dynamics. The open boundary conditions are obtained from the NOAA whole lake circulation model maintained by GLERL. Further modeling work will integrate the particle and biological components. The frequency and intensity of storms is expected to increase in future years, which would correspond to increases in pathogen loads carried by stormwater runoff and combined sewer overflows. This work will be important for water resource managers, regulatory agencies, and urban planners so that they may implement strategies for minimizing pathogen inputs into the Great Lakes.
Products and Services:
- Coupled hydrodynamic/biological model to predict pathogen distribution and fate in urban coastal systems. This model may be used by water resource planners, regulator agencies, and will be useful for prediction of adverse outcomes due to climate change.
- Fecal pollution source tracking tools. This project utilizes a suite of genetic markers, including alternative indicators that will be validated during actual widespread contamination events; e.g. sewage overflows.
Investigator: Bradley S. Moore (UCSD)
Funding Period: October 2007 - September 2008
Region: California Coast
Abstract: Recent fermentation studies have identified actinomycetes of the marine-dwelling genus Salinispora as prolific natural product producers of structurally novel bioactive compounds. To further evaluate their biosynthetic potential, we analyzed all identifiable secondary natural product gene clusters from the recently sequenced 5+ Mbp circular genomes of S. tropica CNB-440 and S. arenicola CNS-205. Our analyses show that their biosynthetic potential meets or exceeds that shown by previous Streptomyces genome sequences as well as other natural product-producing actinomycetes. While a few clusters appear to encode molecules previously identified in Streptomyces species, the majority of the biosynthetic loci are novel. We have employed a genome-inspired strategy to probe the biosynthetic capabilities of these marine bacteria in which we have engineered new chemical entities belonging to the salinosporamide family of potent anticancer agents and have discovered new natural products and novel biochemical pathways. This project has unequivocally demonstrated the potential for genomic analysis to complement and strengthen traditional natural product isolation studies and firmly establishes the genus Salinispora as a rich source of novel drug-like molecules.
Products and Services:
- The 5,183,331-bp genome sequence of Salinispora tropica CNB-440 has been deposited in the GenBank database (accession no. CP000667).
- The partial (33-kb) salinosporamide A biosynthetic sequence of Salinispora tropica CNB-476 has been deposited in the GenBank database (accession no. EF397502).
Investigator: Michael Moore and Rebecca Gast, Woods Hole Oceanographic Institution (WHOI).
Funding Period: October 2005 - September 2009
Region: Gulf of Maine and Continental shelf south to Virginia: NOAA .North Atlantic. region
Abstract: We have surveyed marine mammals and birds (330 cases) in the Northeast United States for evidence of sub-clinical, and clinical zoonoses (disease of risk to humans) in the period October 2005 to September 2007. We are targeting grossly normal and diseased individuals and have developed a molecular screen system that is beginning to be used for other US regions (e.g. strandings in Hawaii).
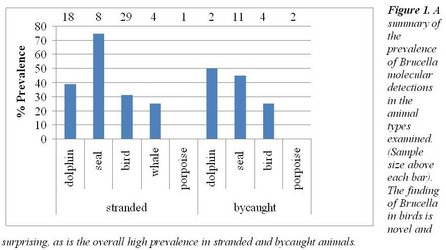 We undertake routine diagnostic necropsy and indicated microbiological culture of stranded and bycaught marine mammal and bird species as practical, sampling gross pathological lesions; also routinely sampling selected normal tissues for microbiological and molecular screening. We screen for the prevalence of influenza, Leptospira, Brucella, Giardia and Cryptosporidium, in addition to routine screens for bacterial infections and their degree of antibiotic resistance. For the targeted zoonoses, avian influenza, Cryptosporidium, Giardia, Brucella and Leptospira were detected. Brucella was the most prevalent. In addition to these analyses of targeted organisms, we also ran routine microbiological cultures on swabs from the thorax, abdomen, respiratory tract and anus/ cloaca. Here we found a broad diversity of organisms known to be either zoonotic or noscomial agents. Noscomial agents are those that affect immunosupressed individuals or are introduced with invasive medical procedures. A surprising level of resistance to multiple antibiotics was also encountered. Our results are of interest to local, regional and national public health agencies as they grapple with the issues to determine where to apply their limited resources. Some of the beaches where we sample scat from marine birds and mammals are heavily used by humans. Having a good baseline survey of the potential risks of humans acquiring infections from these animals is invaluable. Equally important, our data are showing the degree to which microorganisms that have acquired antibiotic resistance in terrestrial treatment regimens, are being circulated throughout the continental shelf ecosystem.
We undertake routine diagnostic necropsy and indicated microbiological culture of stranded and bycaught marine mammal and bird species as practical, sampling gross pathological lesions; also routinely sampling selected normal tissues for microbiological and molecular screening. We screen for the prevalence of influenza, Leptospira, Brucella, Giardia and Cryptosporidium, in addition to routine screens for bacterial infections and their degree of antibiotic resistance. For the targeted zoonoses, avian influenza, Cryptosporidium, Giardia, Brucella and Leptospira were detected. Brucella was the most prevalent. In addition to these analyses of targeted organisms, we also ran routine microbiological cultures on swabs from the thorax, abdomen, respiratory tract and anus/ cloaca. Here we found a broad diversity of organisms known to be either zoonotic or noscomial agents. Noscomial agents are those that affect immunosupressed individuals or are introduced with invasive medical procedures. A surprising level of resistance to multiple antibiotics was also encountered. Our results are of interest to local, regional and national public health agencies as they grapple with the issues to determine where to apply their limited resources. Some of the beaches where we sample scat from marine birds and mammals are heavily used by humans. Having a good baseline survey of the potential risks of humans acquiring infections from these animals is invaluable. Equally important, our data are showing the degree to which microorganisms that have acquired antibiotic resistance in terrestrial treatment regimens, are being circulated throughout the continental shelf ecosystem.
Products and Services:
- Routine gross necropsy of stranded and bycaught marine birds and mammals
- Molecular screening technology for zoonoses that include: influenza, Leptospira, Brucella, Giardia and Cryptosporidium
- Characterization of zoonosis infection prevalences in marine vertebrates for the US Northeast Atlantic Ocean
- Characterization of antibiotic resistance patterns in marine vertebrates for the US Northeast Atlantic Ocean
- Molecular screening for samples submitted from other regions
- Tools for assessing the risk of beach exposure to zoonotic infections
- Six papers published papers and one under review (11/08). We have undertaken numerous public presentations at scientific meetings and in more informal public outreach settings. We have discussed our work with local and regional public health managers, such as the Barnstable County Department of Public Health and the Department of Public Health of the Commonwealth of Massachusetts.
Investigator: Jerome Naar (Center for Marine Science-UNCW, Wilmington NC)
Partners: Fish and Wildlife Research Institute, St. Petersburg, FL.; Food & Drug Administration, Dolphin Island, AL; Florida Department of Health, Tallahassee, FL; US Army, Ft Detrick MD
Funding Period: October 2005 - September 2009
Region: Gulf of Mexico and East Coast
Summary and Highlights: We have recently demonstrated that brevetoxins can accumulate to high levels in live fish, challenging the claim that ichthyotoxicity of brevetoxins precludes accumulation in fish. As such and in the light of the mass mortality of bottlenose dolphin in the Florida Panhandle during spring of 2004, a legitimate public health concern has been raised.
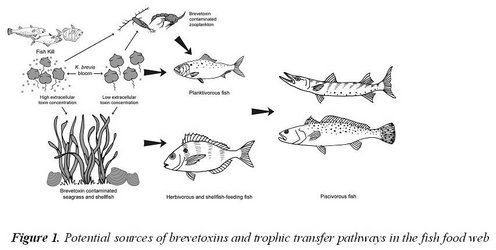 Given the long history of K. brevis blooms in the Gulf of Mexico, and the relatively few, but recent, red tide-related dolphin mortalities, it is not clear whether these mortalities reflect the occurrence of some highly unusual bloom conditions or if they signal an emerging threat regarding brevetoxin accumulation in finfish with obvious implications for human health. Our latest results reveal that, analogous to ciguatoxins, brevetoxins can accumulate in live fish by dietary transfer. Despite the almost annual occurrence of red tide in the eastern Gulf of Mexico, and with the exception of one anecdotal case involving consumption of whole fish (NRC, 1999), human intoxications from eating fish caught during K. brevis red tides have not been documented. In the Florida panhandle, we have observed levels of brevetoxins in the muscle of fish at all trophic levels rise significantly, but not to dangerous levels, during a relatively brief red tide. However, the Florida panhandle is far-less impacted by K. brevis blooms than the southwest coast of Florida, and our findings in that area can not rule out the potential that this could occur. Our current larger-scale monitoring of brevetoxin concentrations in fish muscle from areas in southwest Florida that are more frequently impacted by red tide will help address this issue. Nevertheless, it is clear that brevetoxin concentrations in the liver and digestive tracts of fish can become excessively high and would likely results in acute intoxication if consumed.
Given the long history of K. brevis blooms in the Gulf of Mexico, and the relatively few, but recent, red tide-related dolphin mortalities, it is not clear whether these mortalities reflect the occurrence of some highly unusual bloom conditions or if they signal an emerging threat regarding brevetoxin accumulation in finfish with obvious implications for human health. Our latest results reveal that, analogous to ciguatoxins, brevetoxins can accumulate in live fish by dietary transfer. Despite the almost annual occurrence of red tide in the eastern Gulf of Mexico, and with the exception of one anecdotal case involving consumption of whole fish (NRC, 1999), human intoxications from eating fish caught during K. brevis red tides have not been documented. In the Florida panhandle, we have observed levels of brevetoxins in the muscle of fish at all trophic levels rise significantly, but not to dangerous levels, during a relatively brief red tide. However, the Florida panhandle is far-less impacted by K. brevis blooms than the southwest coast of Florida, and our findings in that area can not rule out the potential that this could occur. Our current larger-scale monitoring of brevetoxin concentrations in fish muscle from areas in southwest Florida that are more frequently impacted by red tide will help address this issue. Nevertheless, it is clear that brevetoxin concentrations in the liver and digestive tracts of fish can become excessively high and would likely results in acute intoxication if consumed.
Products and Services: This project will provide the information needed for health officials to assess whether or not there is a human health risk associated with consumption of fish harvested during or after Karenia brevis blooms. This information will be of prime importance for health officials to take the necessary action regarding monitoring, public education, and prevention programs to be implemented.
Investigator: Joseph Quattro (Univ of South Carolina)
Partners: Hollings Marine Laboratory and Center of Excellence in Oceans and Human Health and NOS/NCOS/CCEHBR
Funding Period: July 2009 - June 2012
Region:
Summary and Highlights: Problem Statement/Objectives: Increased urbanization and utilization of coastal regions has created broad challenges to marine ecosystems and human health. Resultant stressors of augmented land use, such as increased dispersion of toxic chemicals into the marine environment, contribute to the decline of ecosystem health and overall sustainability. Molecular approaches have been used in concert with traditional ecotoxicological data to enhance our understanding of contaminant impact on estuarine communities. Since traditional assessments of environmental health have focused generally on population- and ecosystem-level measurements, integration of ecotoxicological assays combined with molecular approaches will ultimately lead to enhanced understanding of ecosystem health and public safety. We propose to apply modern genomic approaches to create a coastal ecosystem biosensor centered on the marine .sentinel. species P. pugio, an animal especially sensitive to contaminants, that can provide a biological .early warning. system for human health risk by responding early and quickly to environmental challenges. Grass shrimp play an ecologically important role in marine ecosystems, are widely distributed geographically, are easy to rear and manipulate in the laboratory, and have available extensive ecological, toxicological, demographic and genetic information. We suggest that toxicants relevant to coastal ecosystems will produce specific genetic signatures that are valuable biosensors in determining ecosystem health, and, in combination with existing toxicological data, will be useful in the management of estuarine ecosystems.
We propose to generate contaminant specific gene expression profiles in field and laboratory settings using a comprehensive P. pugio microarray, evaluate these arrays as biosensors of ecosystem health, and incorporate gene expression data in ecosystem management plans. We have developed a comprehensive microarray that features ~3,200 partially-annotated genes and exposed male /P. pugio to several contaminants including PBDE and endosulphan at sublethal, environmentally relevant concentrations. Our initial results suggest patterns of gene expression that are both time-course and contaminant specific. We plan to replicate these experiments, include other relevant time-points, contaminants and concentrations (including mixtures), and ultimately assay sex-specific responses. These laboratory exposures will provide baseline data on the impact of commonly encountered contaminants on the P. pugio transcriptome, and provide a linkage between observed transcriptional responses and current measures of toxicity. Our field efforts will establish transcriptional profile baselines for .pristine. environments and make comparisons to .impacted. locations. Site selection will be driven by an ongoing NOAA survey of the population dynamics of P. pugio along coastal South Carolina that has identified populations sequentially impacted by several pesticides over a 15-year period and an on-going complementary study of transcriptional profiling in oyster populations inhabiting adjacent habitats. We will not directly assess contaminant loads, but rather assess departures from transcript profiles in the reference sites and use pre-existing data to gauge .contaminated.. Departures in transcript profiles from reference sites will be viewed as an indication that more in depth surveys including comprehensive contaminant profiles are in order.
Products and Services:
Investigator: Michael Schmale (University of Miami - Cooperative Institute for Marine and Atmospheric Studies)
Partners:
Funding Period: July 2009 - June 2012
Region:
Summary and Highlights: Harmful algal blooms (HAB) have long been a human health concern due to exposure via contamination of food or drinking water, as well as by direct contact (including by inhalation) with a diverse and complex array of toxin molecules. Detection of identified HAB toxins often requires expensive or cumbersome assay systems. In addition, the isolation and identification uncharacterized toxins is often inefficient, particularly with regard to understanding potential adverse health effects on higher organisms.
The goal of the proposed study is to develop and apply tools for detection and analysis of harmful algal HAB toxins using zebrafish (Danio rerio) as a model system. These tools will consist of DNA vectors and transgenic lines of zebrafish containing these vectors. The zebrafish developed here will be useful for toxin detection in high-throughput assays as well as in identification of specific developmental abnormalities in exposed fish. All vectors and transgenic zebrafish lines developed in this study will be made available to government and non-profit agencies and researchers.
The specific aims of the proposed study are:
1. Construct a series of fluorescent protein reporter vectors for HAB toxin detection.
2. Create transgenic zebrafish lines that contain these vectors.
3. Validate the detection efficiency for HAB toxins and determine exposure phenotypes using embryos (eggs) of the transgenic zebrafish.
4. Establish a high-throughput screen for environmental samples using zebrafish eggs in wells of 96-well plates scored in by fluorescent plate reader.
Products and Services:
Investigator: James Oliver(University of North Carolina at Charlotte)
Partners: None
Funding Period: October 2005 - September 2008
Region:
Abstract: Vibrio vulnificus remains the single greatest problem facing the shellfish industry today. Despite being the primary cause seafood-related deaths in this country and an industry-wide recognition of the need to decrease the cases of this pathogen, an overall increase in incidence has occurred during the last 10 years. Fortunately, the number of cases is far less than might be expected based on the number of at-risk persons known to exist in this country. It is possible that one reason for this is that relatively few V. vulnificus strains are able to initiate infection. In support of this hypothesis, we have used DNA sequence data to show that all V. vulnificus strains can be divided into two genetically distinct groups, which we term "C-type" (clinical) and "E-type" (environmental), based on their isolation source. Using a simple and rapid PCR method, we found that 90% of the C-type DNA sequence pattern strains were of clinical origin, and 93% of V. vulnificus strains taken from the environment were the E-type sequence. These differences were statistically highly significant (p < 0.0001). Our findings suggest that C-type strains are relatively rare in the environment, potentially explaining the lower than expected frequency of disease. Although the C-type was rarely isolated from the environment, it was the predominant type isolated from clinical cases. This is an indication that this strain type may be more virulent, and that the C-type genomic pattern could be of value as a strong indicator of virulence potential. Further, our studies suggest a basis for the observation that the ingestion of oysters, even by at-risk consumers, is generally not problematic. The studies we propose are to conduct an extensive evaluation on the occurrence of the two genotypes in the environment, concentrating on their presence in oysters and surrounding waters, and on characterizing the genetics, physiology, and metabolism of the two types. The goal is to identify traits which will allow both their ready detection and quantification, and which may lead to practical methods of determining whether or not the oysters from a given environment would likely contain the more infectious "C-type" strains. To this end, we wished to:
1. Characterize the ecological differences between C and E strains in oysters and surrounding waters.
2. Characterize physiological differences between C and E strains.
3. Examine any geographic differences in C and E strain of V. vulnificus.
Research Highlights: As a summary of our proposal, our lab has now typed the genome of 646 V. vulnificus isolates taken from 75 oysters in both North Carolina and Florida. Surprisingly, we found that of the 646 oyster isolates, only 13.3% were of the C-genotype, whereas 86.7% were of the E-genotype. This difference carries the very high statistical reliability of p < 0.001%. This finding alone may offer some explanation as to why more susceptible oyster-consumers do not develop V. vulnificus infections - numbers of the more infectious C-type of the pathogen may be sufficiently low in most oysters to preclude causing disease. When examined on a seasonal basis, both C- and E-genotype strains of V. vulnificus were observed to increase in both oysters and oyster-growing waters in parallel to water temperature (Fig. 1),
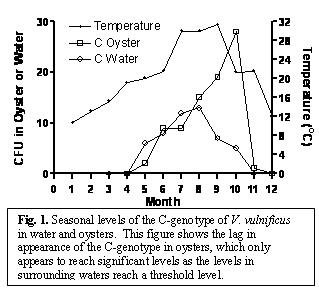
our discovery that the genotype ratio of 129 V. vulnificus isolates taken from the waters of 31 sites surrounding these oysters is not skewed towards E-type strains as seen in oysters, but is almost exactly equal (50.4% E to 49.6% C). This strongly suggested some preferential uptake and/or survival of the E-genotype in oysters as compared to the "virulent" C-genotype. It may be especially significant that only a single oyster (~1.2%) of the 81 individual oysters examined to date had more of the C-genotype cells than of the E-genotype. Indeed, whereas the percentage of .E. type strains typically ranged from 83% - 92%, this one oyster had over 61% of the clinically important C type strain. Such preponderance in one oyster of strains which are presumably more able to cause human infection suggests that only a very occasional oyster presents a human health hazard. This again is consistent with the rarity of human V. vulnificus infections.
In an attempt to understand why C strains predominate in human infections, we studied the survival of the two genotypes in human serum. The results (Fig. 2) clearly sho
w the enhanced ability of C strains to survive under these conditions. We also found that the anti-phagocytic capsule which surrounds the cells and which is critical to the a
bility of this bacterium to cause human disease, is lost by E strains, but retained by C strains, which also helps to explain the predominance of C strain in human disease.
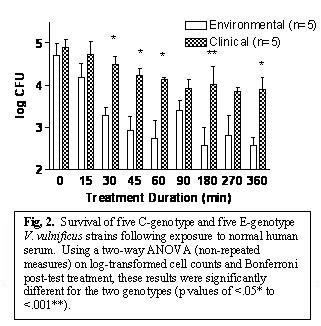
Products and Services: No products, although the results may lead to the development of a rapid method for determining the levels of the two genotypes present in oysters.
Investigator: Mercedes Pascual (University of Michigan)
Partners: Princeton University; Center for Ocean-Land-Atmosphere Studies; University of North Carolina
Funding Period: October 2004 - September 2008
Region: South Asia, Bangladesh
Abstract: One fundamental way in which the oceans are important to human health is through their global role in climate variability. Specifically, Sea Surface Temperature (SST) anomalies (i.e. deviations from their average conditions) might be used to predict the timing and magnitude of epidemic outbreaks for infectious diseases that are climate driven. The feasibility of this approach has been demonstrated by our recent work on endemic cholera in Bangladesh. Sea surface temperature anomalies in the Pacific, an index of the El Niņo Southern Oscillation (ENSO), had been shown in our previous work to influence the incidence of the disease in Matlab and Dhaka, Bangladesh.
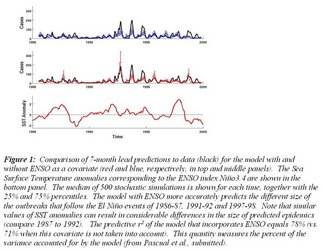
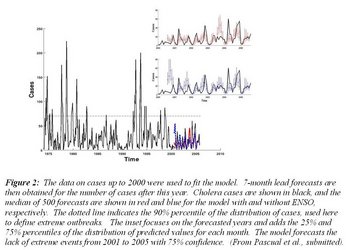 The mathematical/statistical model we had developed to examine this association between climate and disease was more recently put to the test of prediction using the monthly data for the three last decades of cases in Matlab. Hindsight predictive ability was shown to be higher when ENSO (SST anomaly in January) is included in the model (Figure 1). However, this variable alone is not sufficient: the model must also take into account the nonlinear dynamics of the disease because the number of susceptible individuals varies over time as the consequence of previous levels of infection in the population (Figure 1). A more challenging and better test of predictive ability considers the cholera data that had not been used to fit the model. Results show that the model fitted up to year 2000 would have forecasted 7 months ahead with 75% confidence the lack of extreme events and the low disease levels after that year (Figure 2). Parallel efforts for another infectious disease, cutaneous leishmaniasis in Costa Rica, an emergent vector-transmitted disease in the Americas, have also shown the usefulness of time series models that include ENSO for 12-month lead forecasts. These results emphasize the key role of the Oceans in early-warning systems for infectious diseases that are climate sensitive.
The mathematical/statistical model we had developed to examine this association between climate and disease was more recently put to the test of prediction using the monthly data for the three last decades of cases in Matlab. Hindsight predictive ability was shown to be higher when ENSO (SST anomaly in January) is included in the model (Figure 1). However, this variable alone is not sufficient: the model must also take into account the nonlinear dynamics of the disease because the number of susceptible individuals varies over time as the consequence of previous levels of infection in the population (Figure 1). A more challenging and better test of predictive ability considers the cholera data that had not been used to fit the model. Results show that the model fitted up to year 2000 would have forecasted 7 months ahead with 75% confidence the lack of extreme events and the low disease levels after that year (Figure 2). Parallel efforts for another infectious disease, cutaneous leishmaniasis in Costa Rica, an emergent vector-transmitted disease in the Americas, have also shown the usefulness of time series models that include ENSO for 12-month lead forecasts. These results emphasize the key role of the Oceans in early-warning systems for infectious diseases that are climate sensitive.
Products and Services:
- Sofware package written in R for statistical inference of mathematical disease models (under development; available from the authors by request)
- Cholera early-warning system (under development)
- Michael Emch with assistance from the Carolina Population Center Spatial Analysis Unit has finished building a database that is being used to investigate local environmental/climate drivers of cholera in Matlab, Bangladesh and two areas in Vietnam (Hue and Nha Trang). Some of the environmental/climate data are derived from satellites, i.e., sea surface temperature (SST), sea surface height (SSH), and ocean chlorophyll concentration (OCC). Other environmental/climate data are from in situ sources including weather stations and river discharge stations.
Investigator: Mercedes Pascual (University of Michigan)
Partners:
Funding Period: August 2008 - July 2011
Region:
Abstract: This proposal builds on several key elements from our previous project as part of the Oceans and Human Health Initiative:
(1) the quantitative evidence we have amassed for the influence of Pacific sea surface temperature, particularly variations associated with the El Nino-Southern Oscillation, on the dynamics of cholera in Bangladesh;
(2) the understanding that climate drivers alone are not sufficient for prediction and that levels of herd immunity in the population play a key role;
(3) the statistical results linking cholera and precipitation in Bangladesh and the climate model simulations linking Pacific SST to changes in Bangladesh precipitation;
(4) the development of a new and flexible statistical method to fit nonlinear disease models to noisy and partial data;
(5) the initial application of this method to historical disease data which has revealed a different view of cholera immunity and a better mathematical model for the disease ;
(6) our preliminary results on an early-warning system for this disease.
This project brings these individual pieces together in a way that was previously impossible in order to fully develop and validate an early-warning system for endemic cholera. This model will combine our knowledge of climate drivers with the nonlinear dynamics of the disease. We further propose to extend our research from endemic to epidemic dynamics and prediction. The influence of the oceans will be considered at different spatial and temporal scales, from those relevant to El Nino in the Pacific, to those relevant to cholera in the Indian Ocean region, to coastal waters.
Products and Services:
Investigator: Joan Rose and Mantha Phanikumar (Michigan State University)
Funding Period: September 2004 - August 2008
Region: An Integrated Ocean Drilling Program (IODP) expedition to recover sediment samples from the Lomonosov Ridge in the Central Arctic Ocean was successfully executed. Additional sediments for this project were obtained from a hypersaline lake on San Salvador Island, Bahamas, and from coastal environments in the northeast United States.
Abstract: The overwhelming bulk of the biomass inhabiting the world's ocean and its underlying sediments is microbial. While the magnitude of the biomass has been known, we are only beginning to obtain estimates of the genetic diversity that inhabits this enormous realm. In order to better study the microorganisms in the ocean as a biomedical resource, we have engaged a range of techniques to genetically and chemically investigate hitherto unstudied marine microorganisms. A special emphasis is on bacteria derived from the deep subsurface. Microbes dwelling in the deep subsurface are adapted to extreme conditions, including massive pressures, low and high temperatures, and anoxia. We expect that these adaptations may include biosynthetic capabilities unlike those of previously studied microbes. Thus, this untapped reservoir of genetic diversity represents a potential new resource for biomedical research.
Our recent accomplishments include the first investigation of the microbial communities residing under the Arctic seafloor. Prokaryotic DNA was extracted from sediments as deep as 240 meters below the seafloor. Using an array of molecular methods, our results suggest that this environment harbors low bacterial and archaeal biodiversity. Most of the microbes from this extreme environment are likely new to science. In an expedition to the South Pacific Gyre, sediment cores were retrieved from ocean depths of up to 5000 meters. Cultivation of bacteria and culture independent analysis of actinomycete biodiversity from this hitherto uninvestigated environment is currently underway. Additional studies have focused on the discovery of novel antibiotics produced by sediment-derived marine bacteria. A new class of highly potent bisanthraquinone antibiotics has been identified, and semi-synthetic efforts have yielded structure-activity relationships of these drug leads. Antibiotic producing bacteria from hypersaline lake sediments and coastal habitats in the northeast United States have yielded new metabolites that are being evaluated for their chemical and pharmacological properties.
Products and Services:
- The first genetic investigation of prokaryotic communities residing under the Arctic seafloor is contributing to the emerging picture of subsurface life.
- New methods for the acquisition of high quality genetic material from microbes residing in deep subsurface sediments will aid in the future investigation of these extremo philes.
- The first microbiological investigation of Gram-positive bacteria from the South Pacific Gyre will help inform if this remote habitat is a resource for future biomedical discoveries.
- The discovery of structurally novel antibiotics are providing new lead structures for the development of drugs to combat microbial infections.
Investigator: David Senn (Dept. of Environmental Health, Harvard School of Public Health)
Partners: University of Connecticut; Louisiana Universities Marine Consortium (LUMCON)
Funding Period: September 2004 - August 2008
Region: Gulf of Mexico
Abstract: Methylmercury (MeHg) is a neurotoxin that biomagnifies in aquatic foodwebs, and consumption of marine fish and shellfish is the primary MeHg exposure route for humans. In this interdisciplinary study, we are exploring the controls on MeHg production, MeHg biomagnification in foodeweb(s), and human exposure to MeHg in the northern Gulf of Mexico (nGOM) in the Mississippi River influenced area along the Louisiana coast. Despite the socioeconomic importance of the nGOM's recreational and commercial fisheries, mercury (Hg) biogeochemistry and human exposure to MeHg in this system are poorly understood. Over the past 3 years, we undertook 4 research cruises to explore the controls on MeHg production in nGOM sediments, using sediment cores collected along a 400 km east-west transect. We carried out sediment incubation experiments to measure net MeHg production rates, and measured ancillary geochemical parameters to characterize the factors controlling MeHg production in space and time. We also conducted targeted biota collection, focusing on ten key nGOM species of ecological and/or fisheries significance, and measured MeHg levels and indicators of trophic position and food source (stable N and C isotopes) to quantify and explore hypotheses related to MeHg trophic transfer. Finally, we used in-person and internet-based surveys to characterize fish consumption practices of 450 recreational anglers from nGOM, and quantified their MeHg exposure by collecting and measuring Hg in .biomarker. hair samples. Important findings include: i) Hg methylation rates vary by more than a factor 10 in nGOM sediments, and much of this variability can be explained by readily-measured geochemical properties; ii) In fish from the river-influenced area of the nGOM, MeHg levels are strongly associated with .15N (a measure of trophic position; r2 = 0.84) across multiple species and trophic levels of a complex foodweb; ii) MeHg exposure rates are elevated among recreational anglers, with approximately 40% of participants exposed at levels exceeding the US EPA's guideline value of 1 ppm. Data analysis and model development are on-going.
Products and Services
- A mechanistic, geochemically-based, multi-parameter model for identifying key determinants of and predicting net MeHg production in sediments of the nGOM.
- A food-web model for quantitatively characterizing MeHg trophic transfer in fish species of ecological and/or fisheries significance in the nGOM, including the evaluation of the potential interconnectedness of the .green water. (resident) and .blue water. (migratory) fisheries.
- An assessment of MeHg exposure among a .reasonably maximally exposed. subpopulation in the US (i.e., coastal recreational anglers); the identification of major MeHg sources to this subpopulation (e.g., important fish species); and the development and evaluation of survey tools and statistical models for predicting human exposure among similar populations (i.e., what type of information and level of detail is required to accurately assess exposure?).
Investigator: Ramunas Stepanauskas (Bigelow Laboratory for Ocean Sciences)
Partners: Savannah River Ecology Laboratory; Skidaway Institute of Oceanography; J. Craig Venter Institute; University of North Carolina at Charlotte; NOAA Northwest Fisheries Science Center
Funding Period: September 2004 - August 2008
Region: Southeastern U.S.
Abstract: We conducted field surveys, laboratory experiments, and genomic analyses to determine if toxic metal contamination is a significant factor in the proliferation of antibiotic resistance (AR) in coastal water-borne pathogens. Laboratory experiments showed that metal-induced selection for AR was more effective in freshwater compared to seawater, likely due to differences in metal bioavailability at various salinities. We surveyed AR in opportunistic human pathogens Escherichia coli, Vibrio vulnificus, and Vibrio parahaemolyticus in water and sediment samples from one pristine coastal site (the ACE basin, South Carolina) and two metal-contaminated coastal sites (the Shipyard Creek in Charleston, GA, and the LCP Chemical site in Brunswick, GA). We found that, E. coli but not Vibrios had elevated AR in the contaminated sites compared to the ACE basin. Culture-independent analyses demonstrated higher abundance of integrons (genetic elements capable to accumulate multiple genes encoding metal and antibiotic resistances) in the two contaminated sites compared to the ACE basin. However, there was no elevated AR or integron abundance in sediment and ebb tide water samples, compared to flood tide water samples collected in the contaminated creeks. The combined results suggest that AR differences among the sites were caused by large-scale microbial contaminant movement from urban areas rather than by local metal-induced indirect selection.
We found an unexpectedly high frequency of AR in the coastal isolates of E. coli, V. vulnificus, and V. parahaemolyticus. Several strains of E. coli were resistant to record numbers and concentrations of diverse classes of antibiotics, including the first documented cases of resistance to the frontline fluoroquinolones ciprofloxacin and moxifloxacin in environmental E. coli. One of these strains is undergoing whole genome sequencing. Preliminary data suggests that novel fluoroquinolone resistance mechanisms are responsible for this phenomenon, involving multiple efflux pumps. Of particular concern, we found numerous V. vulnificus isolates that were resistant to antibiotics regularly prescribed for V. vulnificus infection treatment, such as doxycyline, tetracycline, aminoglycosides and cephalosporins. V. vulnificus infections are characterized by an extremely short time span between the onset of symptoms and subsequent clinical outcome, and immediate antibiotic therapy for suspected cases is considered critical, highlighting the public health implications of these findings.
Products and Services:
- Our data on antibiotic resistance in V. vulnificus and V. parahaemolyticus may lead to improved treatment of V. vulnificus infections.
- Developed a high-throughput method for antibiotic resistance testing of Vibrio vulnificus, V. parahaemolyticus, and E.coli in coastal monitoring.
- Libraries of 436 E. coli, 157 V. vulnificus, and 696 V. parahaemolyticus isolates were created and are available to the research community as a long-term asset for continuing studies.
- The genome of a multi-resistant E. coli strains is being sequenced to serve as reference material in antibiotic resistance research.
- Generated recommendations for improved V. vulnificus quantification methodology.
Investigator: Peter Strutton (College of Oceanic and Atmospheric Sciences, Oregon State University)
Partners: University of Oregon
Region: Oregon Coast
Abstract:
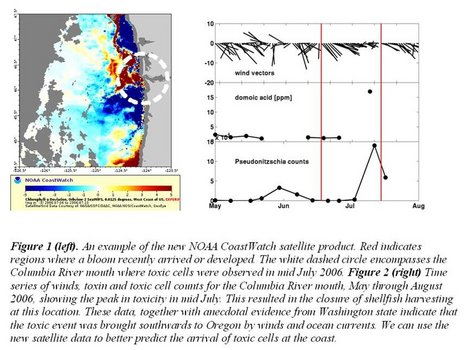 Relevance to OHH and value to society: We are working towards an early warning system that will help protect the public from exposure to algal toxins. The toxins we are primarily interested in are domoic acid, produced by the diatom genus Pseudonitzschia, which leads to amnesic shellfish poisoning and saxitoxin, produced by the dinoflagellate genus Alexandrium, which leads to paralytic shellfish poisoning. Despite the lack of a definitive optical signal for either of these organisms, we are developing ways to use satellite ocean color imagery to identify and track blooms. These techniques will be used to warn coastal managers of the potential landfall of toxic blooms, thus minimizing their impact on consumers of coastal shellfish.
Relevance to OHH and value to society: We are working towards an early warning system that will help protect the public from exposure to algal toxins. The toxins we are primarily interested in are domoic acid, produced by the diatom genus Pseudonitzschia, which leads to amnesic shellfish poisoning and saxitoxin, produced by the dinoflagellate genus Alexandrium, which leads to paralytic shellfish poisoning. Despite the lack of a definitive optical signal for either of these organisms, we are developing ways to use satellite ocean color imagery to identify and track blooms. These techniques will be used to warn coastal managers of the potential landfall of toxic blooms, thus minimizing their impact on consumers of coastal shellfish.
Accomplishments to date: Using historical data on the levels of toxins in shellfish at approximately 20 locations along the Oregon coast, we have identified hot spots of frequent contamination. We have quantified a link between El Niņo events and saxitoxin occurrence. Cluster analysis has identified regions of the coast that are subject to shellfish closures at the same time (or not), thus potentially streamlining the process of identifying .at risk. locations. A new satellite product for bloom identification has been developed and disseminated. Satellite analyses have provided insight into the relationship between bloom development, nearshore winds and landfall of toxic blooms.
Who will or is using your products/services: The Oregon Department of Agriculture is the agency currently responsible for the closure of shellfish beds when toxin concentrations in shellfish exceed a predetermined limit. Our OHH-funded work will be incorporated into a new MERHAB (Monitoring and Event Response for Harmful Algal Blooms) program which coordinates the efforts of state, federal and academic institutions that are addressing various aspects of the HAB problem. Thus, our research findings will contribute to a more sophisticated procedures for determining the closure of shellfish beds to harvesting.
Products and Services:
- New satellite data product for harmful algal bloom detection, the chlorophyll anomaly (or deviation) product, disseminated by the CoastWatch west coast regional node: http://coastwatch.pfeg.noaa.gov/coastwatch/CWBrowser.jsp
- New web site that collates data from the Oregon agencies that monitor shellfish toxins and presents them in an oceanographic context: http://www.coas.oregonstate.edu/habs
Investigator: Gordon T. Taylor (School of Marine and Atmospheric Sciences Stony Brook University)
Funding Period: September 2004 - August 2009
Region: North Atlantic region, specifically Long Island Sound which is representative of estuarine environments generally.
Abstract: The primary goal of this project is to improve our understanding of oceanographic processes that contribute to viral decay and inactivation in coastal waters.
 These processes are obviously relevant to human health concerns, such as safe bathing waters and quality of harvested marine living resource. Because most previous research focused on inactivating effects of light, or within illuminated waters, and most coastal discharges of human waste are into turbid waters, our project focuses on the .dark processes., including physical, chemical and biological interactions. The project combines laboratory experimentation with field collections from sites heavily impacted by sewage discharge as well as more pristine reference sites. Our laboratory experiments demonstrate that responses may vary among bacteriophages. For T4, marine particles between 0.2 and 10 .m appear more important in viral inactivation than dissolved constituents. Removal or alteration of dissolved constituents (< 0.2 .m) by autoclaving, ultrafiltration and UV-oxidation did little to improve viral survival. beyond particle removal (Figure and Table below). For the marine P1 phage, heat-labile dissolved constituents were as important as particles. Paradoxically, viral inactivation was inversely related to concentrations of dead plankton debris in seawater seeded with T4 bacteriophage and varying amounts of autoclaved diatom culture suspension. In toto, our results suggest that organisms in the 0.2-10 .m size fraction and heat-labile solutes are primarily responsible for viral inactivation and disappearance. We tested whether sorption to living non-competent bacterial cells (non-host species and resistant strain of host species) or enzymatic attack may be the cause of viral inactivation. We found little evidence to support these hypotheses using a single model virus and two marine bacterial strains.
These processes are obviously relevant to human health concerns, such as safe bathing waters and quality of harvested marine living resource. Because most previous research focused on inactivating effects of light, or within illuminated waters, and most coastal discharges of human waste are into turbid waters, our project focuses on the .dark processes., including physical, chemical and biological interactions. The project combines laboratory experimentation with field collections from sites heavily impacted by sewage discharge as well as more pristine reference sites. Our laboratory experiments demonstrate that responses may vary among bacteriophages. For T4, marine particles between 0.2 and 10 .m appear more important in viral inactivation than dissolved constituents. Removal or alteration of dissolved constituents (< 0.2 .m) by autoclaving, ultrafiltration and UV-oxidation did little to improve viral survival. beyond particle removal (Figure and Table below). For the marine P1 phage, heat-labile dissolved constituents were as important as particles. Paradoxically, viral inactivation was inversely related to concentrations of dead plankton debris in seawater seeded with T4 bacteriophage and varying amounts of autoclaved diatom culture suspension. In toto, our results suggest that organisms in the 0.2-10 .m size fraction and heat-labile solutes are primarily responsible for viral inactivation and disappearance. We tested whether sorption to living non-competent bacterial cells (non-host species and resistant strain of host species) or enzymatic attack may be the cause of viral inactivation. We found little evidence to support these hypotheses using a single model virus and two marine bacterial strains.
Field-collected viral concentrates from near sewage outfalls and more pristine waters have been used in mammalian tissue culture challenges as well as quantitative RT-PCR. Analyses of these samples and data are ongoing.
Products and Services:
- Information is the primary product from this project. Better understanding of viral inactivation in coastal waters, and successful modification of Q-PCR and infectivity assays, are anticipated to lead to a standardized set of protocols that will be useful in unequivocally determining coastal water quality with respect to human pathogenic viruses. These tools will be useful for coastal zone managers, the academic community, monitoring agencies, and possibly the Office of Homeland Security. Modeling and forecasting products from this project will contribute to development of better models of viral dynamics in coastal waters and thereby contribute to better forecasting of viral responses and management of coastal resources.
Investigator: Catherine J. Walsh (Mote Marine Laboratory, Sarasota, FL)
Partners: University of South Florida/All Children's Hospital, St. Petersburg, FL; Centers for Disease Control and Prevention, Atlanta, GA
Funding Period: October 2005 - September 2009
Region: The geographic scope of this project includes southwestern Florida where blooms of Karenia brevis are common. Laboratory experimentation is being conducted at Mote Marine Laboratory in Sarasota, FL and at the University of South Florida in St. Petersburg, FL. Air sampling measurements and collection of nasal secretions and blood samples from occupationally exposed healthy volunteers occurs on Siesta Key Beach in Sarasota, FL. Since the geographic focus of this project is in Florida, the project encompasses two NOAA regions, the Gulf of Mexico and Southeast & Carribean.
Abstract: The toxic dinoflagellate, Karenia brevis, produces a suite of polyether neurotoxins, termed brevetoxins, which result in severe respiratory effects in humans.
 Accumulating evidence suggests initial respiratory effects may represent only part of the overall consequences associated with toxin exposure, and chronic health implications have not been well studied. Understanding effects of marine biotoxins on human health is one of the priority areas described in NOAA's Oceans and Human Health Initiative. By focusing on contributing to the current understanding of potential impacts of brevetoxins on human health, particularly immune function, this project is directly responsive to this goal. To date, major research findings indicate that in vitro exposure of immune cells to brevetoxins results in programmed cell death, or apoptosis. The toxin congeners PbTx-2 and PbTx-6, but not PbTx-3, induce apoptosis in a T lymphocyte cell line as well as in human primary peripheral blood mononuclear cells (PBMC). Proliferation of a T lymphocyte cell line as well as PHA-stimulated human PBMC was decreased in the presence of PbTx-2 or PbTx-6, but not PbTx-3. Using a monocyte cell line, cellular metabolism of PbTx-2 was documented by the formation of two brevetoxin conjugates, including a cysteine conjugate and a conjugate formed with the intracellular thiol, glutathione. Cytokines in nasal secretions from occupational or volunteer exposure to brevetoxins did not demonstrate detectable changes in cytokine profiles. Results from this research will be useful to other scientists conducting research related to human health effects associated with red tide toxins. Public health partners including the Florida Department of Health and Centers for Disease Control and Prevention will also benefit from project results. Characterization of cellular consequences of brevetoxin exposure is critical to fully understand the impact of recurrent red tide events on human health and to reduce potential health consequences associated with red tide toxin exposure.
Accumulating evidence suggests initial respiratory effects may represent only part of the overall consequences associated with toxin exposure, and chronic health implications have not been well studied. Understanding effects of marine biotoxins on human health is one of the priority areas described in NOAA's Oceans and Human Health Initiative. By focusing on contributing to the current understanding of potential impacts of brevetoxins on human health, particularly immune function, this project is directly responsive to this goal. To date, major research findings indicate that in vitro exposure of immune cells to brevetoxins results in programmed cell death, or apoptosis. The toxin congeners PbTx-2 and PbTx-6, but not PbTx-3, induce apoptosis in a T lymphocyte cell line as well as in human primary peripheral blood mononuclear cells (PBMC). Proliferation of a T lymphocyte cell line as well as PHA-stimulated human PBMC was decreased in the presence of PbTx-2 or PbTx-6, but not PbTx-3. Using a monocyte cell line, cellular metabolism of PbTx-2 was documented by the formation of two brevetoxin conjugates, including a cysteine conjugate and a conjugate formed with the intracellular thiol, glutathione. Cytokines in nasal secretions from occupational or volunteer exposure to brevetoxins did not demonstrate detectable changes in cytokine profiles. Results from this research will be useful to other scientists conducting research related to human health effects associated with red tide toxins. Public health partners including the Florida Department of Health and Centers for Disease Control and Prevention will also benefit from project results. Characterization of cellular consequences of brevetoxin exposure is critical to fully understand the impact of recurrent red tide events on human health and to reduce potential health consequences associated with red tide toxin exposure.
Products and Services:
- Scientific manuscript on apoptosis and proliferation in a T lymphocyte cell line. In review, available in approximately 3 months.
- Scientific manuscript on apoptosis and cellular proliferation in human primary immune cells. In preparation, available in approximately 3- 6 months.
- Scientific manuscript on cellular metabolism of toxins. In preparation, available 3-6 mos.
- Database on cDNA microarray of human primary PBMC treated with brevetoxins (PbTx-2) in vitro. In preparation. Available 3-6 months.
Investigator: Guangyi Wang (Department of Oceanography, University of Hawaii)
Funding Period: September 2004 - August 2008
Region: Pacific islands . Reef Ecosystems of the 8 main Hawaiian Islands. Results of this project are also important in the study of the impact of marine fungi on oceans and human health in other regions.
Abstract: Marine fungi are well established as a significant source for novel pharmaceutical compounds. Some fungi are disease-causing agents and others produce toxin.
 Still others are pathogens to both immune-compromised marine animals and beach-swimmers. The proposed research is to investigate diversity of marine fungi in the Hawaiian Islands and screen the isolated marine fungal isolates for bioactivities. As such, the proposed work is closely relevant to OHH by providing information on ocean-related health threats from fungal pathogens and new fungal strains for the development of novel pharmaceutical compounds.
Still others are pathogens to both immune-compromised marine animals and beach-swimmers. The proposed research is to investigate diversity of marine fungi in the Hawaiian Islands and screen the isolated marine fungal isolates for bioactivities. As such, the proposed work is closely relevant to OHH by providing information on ocean-related health threats from fungal pathogens and new fungal strains for the development of novel pharmaceutical compounds.
The major research findings and accomplishments to date include: 1). Collection of near one thousand of marine samples including draft woods, algae, mangrove, and sponges have been collected from 7 major Hawaiian islands; 2) establishment of Hawaii Marine Fungal Collection (HMFC) containing over 1,500 fungal strains, including over 200 new species; 3) identification of over 100 fungal strains producing interesting bioactivities; 4) identification of unexpected fungal diversity and fungal pathogens from marine species (alien sponges and algae) and other substrates; 5) identification of unexpected fungal pathogens with potential ability to produce toxins; and 6) Establishment of rDNA-ITS database for barcoding marine fungi.
The findings of this project allow the scientific community and the general public to gain knowledge of fungal diversity and to be familiar with the potential risks associated with fungal pathogens along the coasts of the Hawaii Islands. Fungal diversity information and fungal strains resulted from this project are extremely valuable for the scientific community. Particularly, new fungal strains can be used for research on novel natural compounds and industrial enzymes. Finally, results of the proposed research will be very useful to guide decision making for further investigation of coastal regions of US mainland and coastal ecosystem management in Hawaii.
Products and Services:
- 1. Hawaiian Marine Fungal Collection (HMFC)-contains over 600 marine fungal stains;
- 2. Culturable fungal stain data-including collection sites, genus or species name, and rDNA-ITS sequences;
- 3. Fungal strain distribution - providing fungal stains for the natural compound structure identification to researchers in Departments of Chemistry at University of Hawaii (Philip Williams) and Hawaii Pacific University (David Horgen).
Investigator: Philip J. W. Roberts (Georgia Tech. University)
Funding Period: April 2006 - April 2008
Region: The particular region under study is the nearshore of Lake Michigan near Grand Haven that is impacted by the Grand River plume. The models under development are applicable to more general lake and marine regions impacted by river-borne pollutants, however.
Abstract: The fate and transport of bacterial contaminants carried by river plumes entering lakes and coastal waters is being studied.
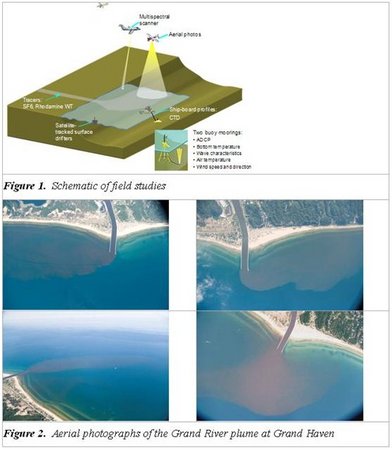 The ultimate objective is to develop mathematical models of plume dynamics as part of the NOAA OHHI research goal to develop early warning systems for health risks. The plumes can carry pathogens due to surface runoff and, more seriously, combined sewer overflows (CSO's). These pathogens pose serious human health risks by impinging on local beaches, potentially closing them, and by entrainment into water intakes. Four comprehensive field experiments (Figure 1) have been completed on the plume from the Grand River, which is subject to CSO's from Grand Rapids, as it enters Lake Michigan at Grand Haven (Figure 2). These experiments include aerial photography and simultaneous measurements of lake physical properties. The spatial extent of the plume is measured by traverses of salinity, temperature, and depth from a boat. The river water is generally warmer than the lake waters, causing the plume to form a thin surface layer about one to two meters thick that spreads dynamically. Preliminary findings show the plume behavior to be complex, and highly variable, as in the Figure 2 examples. Its shape and direction is highly dependent on lake currents and winds and its mixing dependort of pathogens to local recreation areas. We are using the three-dimensional model developed by GLERL to drive a high-resolution grid model around Grand Haven. Even this grid is too coarse to properly model the small-scale river plume dynamics; however, so a separate model is being developed that will incorporate the correct plume physics, such as buoyant surface spreading, into the gridded model.
The ultimate objective is to develop mathematical models of plume dynamics as part of the NOAA OHHI research goal to develop early warning systems for health risks. The plumes can carry pathogens due to surface runoff and, more seriously, combined sewer overflows (CSO's). These pathogens pose serious human health risks by impinging on local beaches, potentially closing them, and by entrainment into water intakes. Four comprehensive field experiments (Figure 1) have been completed on the plume from the Grand River, which is subject to CSO's from Grand Rapids, as it enters Lake Michigan at Grand Haven (Figure 2). These experiments include aerial photography and simultaneous measurements of lake physical properties. The spatial extent of the plume is measured by traverses of salinity, temperature, and depth from a boat. The river water is generally warmer than the lake waters, causing the plume to form a thin surface layer about one to two meters thick that spreads dynamically. Preliminary findings show the plume behavior to be complex, and highly variable, as in the Figure 2 examples. Its shape and direction is highly dependent on lake currents and winds and its mixing dependort of pathogens to local recreation areas. We are using the three-dimensional model developed by GLERL to drive a high-resolution grid model around Grand Haven. Even this grid is too coarse to properly model the small-scale river plume dynamics; however, so a separate model is being developed that will incorporate the correct plume physics, such as buoyant surface spreading, into the gridded model.
Products and Services:
- The major product/service from this project will be a reliable mathematical model that will be part of an early-warning system for bacterial beach contamination.
- The scientific contribution is improved understanding of complex dynamics of river plumes and their behavior under various forcing mechanisms.
- Methods to predict plume entrainment into water intakes.
Investigator: David C. Rowley (College of Pharmacy, University of Rhode Island)
Funding Period: September 2004 - August 2008
Region:
 An Integrated Ocean Drilling Program (IODP) expedition to recover sediment samples from the Lomonosov Ridge in the Central Arctic Ocean was successfully executed. Sediment samples for genetic and cultivation experiments were collected from two sites (87.92°N, 139.37° W and 87.87°N, 138.18° W), both with a water column depth of >1200 meters. Core samples were retrieved from depths of 12 - 348 meters below the sea floor. During a recent (Dec 06 - Jan 07) expedition to the South Pacific Gyre, eleven sites (figure at right) were cored to a maximum depth of .18 m below sea floor. Additional sediments for this project were obtained from a hypersaline lake on San Salvador Island, Bahamas, and from coastal environments in the northeast United States.
An Integrated Ocean Drilling Program (IODP) expedition to recover sediment samples from the Lomonosov Ridge in the Central Arctic Ocean was successfully executed. Sediment samples for genetic and cultivation experiments were collected from two sites (87.92°N, 139.37° W and 87.87°N, 138.18° W), both with a water column depth of >1200 meters. Core samples were retrieved from depths of 12 - 348 meters below the sea floor. During a recent (Dec 06 - Jan 07) expedition to the South Pacific Gyre, eleven sites (figure at right) were cored to a maximum depth of .18 m below sea floor. Additional sediments for this project were obtained from a hypersaline lake on San Salvador Island, Bahamas, and from coastal environments in the northeast United States.
Abstract: The overwhelming bulk of the biomass inhabiting the world.s ocean and its underlying sediments is microbial. While the magnitude of the biomass has been known, we are only beginning to obtain estimates of the genetic diversity that inhabits this enormous realm. In order to better study the microorganisms in the ocean as a biomedical resource, we have engaged a range of techniques to genetically and chemically investigate hitherto unstudied marine microorganisms. A special emphasis is on bacteria derived from the deep subsurface. Microbes dwelling in the deep subsurface are adapted to extreme conditions, including massive pressures, low and high temperatures, and anoxia. We expect that these adaptations may include biosynthetic capabilities unlike those of previously studied microbes. Thus, this untapped reservoir of genetic diversity represents a potential new resource for biomedical research.
Our recent accomplishments include the first investigation of the microbial communities residing under the Arctic seafloor. Prokaryotic DNA was extracted from sediments as deep as 240 meters below the seafloor. Using an array of molecular methods, our results suggest that this environment harbors low bacterial and archaeal biodiversity. Most of the microbes from this extreme environment are likely new to science. In an expedition to the South Pacific Gyre, sediment cores were retrieved from ocean depths of up to 5000 meters. Cultivation of bacteria and culture independent analysis of actinomycete biodiversity from this hitherto uninvestigated environment is currently underway. Additional studies have focused on the discovery of novel antibiotics produced by sediment-derived marine bacteria. A new class of highly potent bisanthraquinone antibiotics has been identified, and semi-synthetic efforts have yielded structure-activity relationships of these drug leads. Antibiotic producing bacteria from hypersaline lake sediments and coastal habitats in the northeast United States have yielded new metabolites that are being evaluated for their chemical and pharmacological properties.
Products and Services:
- The first genetic investigation of prokaryotic communities residing under the Arctic seafloor is contributing to the emerging picture of subsurface life.
- New methods for the acquisition of high quality genetic material from microbes residing in deep subsurface sediments will aid in the future investigation of these extremophiles.
- The first microbiological investigation of Gram-positive bacteria from the South Pacific Gyre will help inform if this remote habitat is a resource for future biomedical discoveries.
- The discovery of structurally novel antibiotics are providing new lead structures for the development of drugs to combat microbial infections.
Investigator: Cheryl A. Cross (University of Tennessee, College of Veterinary Medicine)
Partners: Center for Coastal Environmental Health and Biomolecular Research (CCEHBR)
Funding Period: June 2006 - September 2008
Region: Western and Southeastern U.S. and Caribbean, Gulf of Mexico
Abstract: Current research investigates the systemic effects and pathology of experimental domoic acid (DA) toxicity, an algal biotoxin and analogue of the neurotransmitter glutamate which causes morbidity and mortality in marine life and humans ingesting contaminated seafood.
Microscopic evaluation is performed to compare experimental pathology with natural exposure cases, and attempt prophylactic therapies that may ameliorate disease. Microscopic investigation thus far shows excellent correlation between experimental and natural disease, with the primary brain target region being the limbic system.
Upregulation of the inflammatory mediator cyclooxygenase 2 (COX-2) following experimental exposure and prophylactic use of COX-2 inhibitors is being tracked using immunohistochemistry (antibody-linked stain which detects an antigen in tissue section) in rodents. Findings thus far suggest COX-2 is upregulated in the limbic system, and that administration of selective and nonselective COX-2 inhibitors decrease morbidity, mortality, and microscopic pathology (including neuronal degeneration and an increase in number and size of astrocytes).
This is the first known experimental research showing a cardiac lesion following DA exposure that is microscopically similar to that described in California sea lions (CSL) and sea otters. Immunohistochemistry for anti-active caspase 3 (a marker of apoptosis, or programmed cell death) is being applied to DA-associated heart lesions of exposed mice, CSL's, and sea otters. Results thus far in mice and CSL suggest the lesion is predominantly apoptotic and not necrotic, suggesting an oxidant injury, thus allowing targeted therapeutic intervention.
Planned experiments for the next year include tracking neural migration patterns following experimental (rodents) and natural (CSL) in utero DA exposure, and further classifying the mitigation of toxicity with the prophylactic use of COX-2 inhibitors.
The project goals are to link experimental findings with those of natural exposure cases to better classify DA toxicity, investigate potential therapies following exposure, and decrease the morbidity and mortality associated with the toxin.
Products and Services:
- Routine necropsy
- Tissue processing, slide production, routine and special section staining
- Immunohistochemistry for COX-2, GFAP (astrocyte marker), and anti-active caspase 3 (apoptosis marker)
- Immunohistochemistry for mature neurons, neural stem cells, and premature neurons and tracking neural development following DA exposure (mice and CSL) (proposed)

